Current Research
A Digest of Recent Publications
A complete bibliography is available here.
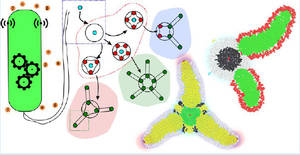 | Jonathan Pascalie, Martin Potier, Taras Kowaliw, Jean-Louis Giavitto, Olivier Michel, Antoine Spicher, and René Doursat. Developmental Design of Synthetic Bacterial Architectures by Morphogenetic Engineering in ACS Synthetic Biology, May, 2016 |
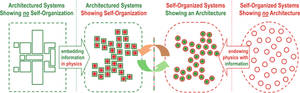 | René Doursat, Hiroki Sayama, and Olivier Michel. A review of morphogenetic engineering in Natural Computing, September, pp 1–19, 2013 |
 | Martin Potier, Antoine Spicher, and Olivier Michel. Topological Computation of Activity Regions in ACM SIGSIM Conference on Principles of Advanced Discrete Simulation (PADS), Montréal, Canada, May 2013 |
 | Antoine Spicher, Olivier Michel, and Jean-Louis Giavitto. Interaction-Based Modeling of Morphogenesis. Chapter in the book Morphogenetic Engineering, Doursat, René; Sayama, Hiroki; Michel, Olivier (Eds.). Dec 2012 |
 | Antoine Spicher, Olivier Michel, and Jean-Louis Giavitto. Arbitrary nesting of spatial computing. In Spatial Computing Workshop 2012 at AAMAS, Valencia, Spain, June 2012. |
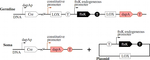 | Antoine Spicher, Olivier Michel, and Jean-Louis Giavitto. Interaction-based simulations for Integrative Spatial Systems Biology. chapter in the book Understanding the dynamics of biological systems, Werner Dubitzky, Jennifer Southgate and Hendrik Fuß editors, 2011, 195-231, pdf file |
 | Louis Bigo, Antoine Spicher and Olivier Michel. Spatial Programming for Music Representation and Analysis. SCW 2010 - Spatial Computing Workshop, 2010, pdf file |
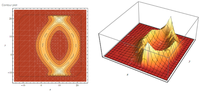 | Alexandre Muzy, Luc Touraille, Hans Vangheluwe, Olivier Michel, Mamadou Kaba Traoré, David R.C. Hill. Activity Regions for the Specification of Discrete Event Systems. DEVS 2010, Symposium on Theory of Modeling and Simulation, 2010. pdf file |
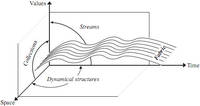 | Antoine Spicher, Olivier Michel and Jean-Louis Giavitto. Spatial Computing as Intensional Data Parallelism. Third IEEE International Conference on Self-Adaptive and Self-Organizing Systems - Spatial Computing Workshop, 2009.pdf file |
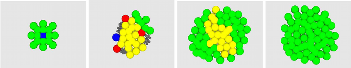 | Olivier Michel, Antoine Spicher, and Jean-Louis Giavitto. Rule-Based Programming for Integrative Biological Modeling. Natural Computing, Volume 8, Issue 4 (2009), Page 865.pdf file1) |
 | Jean-Louis Giavitto, Olivier Michel, and Antoine Spicher. Spatial Organization of the Chemical Paradigm and the Specification of Autonomic Systems. Software-Intensive Systems, pages 235-254, 2008, LNCS 5380 pdf file and at Springer's |
 | Antoine Spicher, Olivier Michel, Mikolaj Cieslak, Jean-Louis Giavitto, and Przemyslaw Prusinkiewicz. Stochastic P systems and the simulation of biochemical processes with dynamic compartments. BioSystems, 91(3):458-472, March 2008. pdf file |
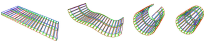 | Antoine Spicher and Olivier Michel. Declarative modeling of a neurulation-like process. BioSystems, 87:281-288, February 2006 pdf file |
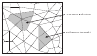 | Antoine Spicher and Olivier Michel. Using rewriting techniques in the simulation of dynamical systems: Application to the modeling of sperm crawling. In Fifth International Conference on Computational Science (ICCS'05), volume I, pages 820-827, 2005 pdf file |
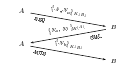 | Olivier Michel and Florent Jacquemard. An Analysis of a Public-Key Protocol with Membranes, pages 283-302. Natural Computing Series. Springer Verlag, 2005. pdf file |
 | Jean-Louis Giavitto, Olivier Michel, Julien Cohen, and Antoine Spicher. Computation in space and space in computation. In J.-P Banâtre, P. Fradet, J.-L. Giavitto, and O. Michel, editors, Unconventional Programming Paradigms (UPP'04), volume 3566 of LNCS, pages 137-152. ERCIM- NSF, Springer Verlag, 2005. pdf file |
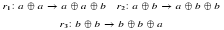 | Jean-Louis Giavitto, Grant Malcolm, and Olivier Michel. Rewriting systems and the modelling of biological systems. Comparative and Functional Genomics, 5:95-99, February 2004. pdf file |
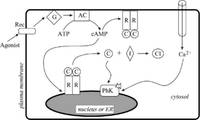 | Jean-Louis Giavitto and Olivier Michel, Modeling the topological organization of cellular processes, BioSystems, 70:149-163, 2003. pdf file |
Context of the project
My current research activities take place in the MGS project. This project evolves following two complementary directions:
- to study and develop the integration of topological notions and tools in programming languages;
- to apply those notions and tools to the conception and development of new data- and control-structures that are expressive and efficient for the modeling and simulation of dynamical systems with a dynamic structure.
This research is concretized by the development of an eponymous experimental programming language and its application to the modeling and simulation of dynamical systems. We have a particular focus in the field of biology and morphogenesis.
Internships
We have proposed some internships on these topics in 2004, 2005, and 2008. Feel free to contact me if you would like to work on this topic. Whatever your background is (physics, biology, mathematics, computer science, etc.) we can always manage to find you a suitable subject.
High-level Domain Specific Programming Languages
My research work in that domain focuses on the definition of new data- and control- structures allowing the representation of spatial and temporal data for the modeling and simulation of dynamical systems in a rigorous framework, close to the end-user and the mathematical tools used in that domain (dynamical systems in biology, chemistry and physics).
The two main applications domains that have motivated the development of these new mechanisms are the analysis and representation of complex relations (for example spatial ones) and the simulation of dynamical systems where the structure has to be computed jointly with the evolution of the system (as it is the case in developmental biology, at any level from the cells to the individuals).
A privileged application domain was the modeling and simulation in systemic biology (more specifically in post-genomic and in integrative simulation of developmental processes). We are now (since 2007) considering the modeling, simulation and conception of new computational resources brought by nanosciences and molecular biology.
This research is based on the development of a new computation paradigm (by generalizing the approaches of the chemical computing, membrane computing, Lindenmayer systems and cellular automatons) and new tools for the interpretation, typing and efficient compilation of declarative languages. This research meets the fields of the “self-*” (self-sustaining, self-healing, self-organizing) systems that are emerging in the software community and whose long-term goal is to identify and develop in today’s computers the fundamental requirements to ensure properties of flexibility, adaptivity, robustness, self-healing that are common to the complex systems of living matter.
The MGS Project
 Everything that concerns MGS and spatial computing can be found in its own webpage. Many examples are given in the image gallery and the interpreter can be found here.
Everything that concerns MGS and spatial computing can be found in its own webpage. Many examples are given in the image gallery and the interpreter can be found here.
 MGS has been used as a modelling language for the Paris team of the MIT iGEM's 2007 competition. The source code of the models can be run with the current version of the MGS interpreter.
MGS has been used as a modelling language for the Paris team of the MIT iGEM's 2007 competition. The source code of the models can be run with the current version of the MGS interpreter.
The french project is targeted to the construction of a multicellular bacterial organism made of two co-existing cell types. Brainstorming resulted in proposing a system made of so-called soma cells, that produce a metabolite (DAP), and will not be able to divide, and of so-called germ cells that are able to grow, but only in presence of (sufficient quantities of) DAP, and are able to differentiate into soma cells. Informal reasoning indicate that this design should be correct, in the sense that it leads to an exponential growth of the two coexisting cell types. However, before actually constructing this system, the design has been assessed using several models at different levels mainly implemented in MGS.
Conferences and Working Groups, Networks...
Membre du GDRp GPL, du GDRS ÉcoInfo et du collectif Labo 1.5.
Recent Presentation in Computing With Respect to the Planetary Boundaries
2025
- 2025 (30/06/2025) présentation Bilan carbone pour laboratoire de recherche à la journée du LACL avec Maëlle Chamault de la DT de l'université
- 2025 (26-27/06/2025) 11th Workshop on Computing Within Limits avec la présentation ICT Within LIMITS is Bound to be Old-Fashioned by Design avec Émilie Frenkiel
2024
- 2024 (03/12/2024) 1st International Workshop on Low Carbon Computing avec la présentation SlowTech LIFE project: second-hand computing devices under intermittent energy supply
- 2024 (15/11/2024) Regards croisés sur les limites planétaires (environnement, droit, numérique) à l'IMAG de Grenoble dans le cadre du projet Aldiwo avec une présentation intitulée Quelles possibilités (de l’enseignement) d'un numérique dans le cadre des frontières planétaires ? où il était question(entres autres) du projet SlowTech
- 2024 (4/11/2024) Penser la soutenabilité environnementale en recherche à l'IPGP avec une présentation intitulée Que(l)(q)les possibilités d'un numérique dans le cadre des limites planétaires
- 2024 (17/10/2024) Colloque/Formation: Comprendre le(s) rôle(s) de l’Intelligence Artificielle générative à l’université avec une présentation intitulée L’IA, d’une informatique sans limites à un usage (ir)responsable ?
- 2024 (6-7/06/2024) International Conference Socioecos. Climate Change, Sustainability and Socio-ecological Practices présentation avec Émilie Frenkiel de Student Citizens’ Assemblies et la publication Student Citizens’ Assemblies, Politics, Sustainability and Socio-ecological Practices
- 2024 (23/05/2024) Journées du laboratoire Matières et Systèmes Complexes de l'Université Paris Cité, présentation Que faire de cet obscur objet du désir ? ou De la limite considérée comme un des beaux arts
- 2024 (09/04/2024) Présentation avec Émilie Frenkiel à l'AFNIC du projet social/écologique/pédagogique/scientifique SlowTech émanant de la CCE et de son positionnement dans les frontières planétaires
- 2023 (31/05/23) Enseigner les conséquences environnementales du numérique au Gdt Politiques environnementales du numérique du GDR Internet, IA et Société
- présentation Enseigner les conséquences environnementales du numérique avec Hajar El Karmouni et Alexandre Brenner (directeur du département SI de l'EPISEN)
- présentation CCE 2022 - Quel numérique pour quelle société avec Émilie Frenkiel
- 2023 (04-05/07/2023) Enseigner les Transitions Écologiques et Sociales dans le supérieur
- présentation Retour d'expérience : une formation ingénieur en informatique à l'aune du paradigme des low-techs avec Hajar El Karmouni et Alexandre Brenner (directeur du département SI de l'EPISEN)
Conferences
- 2013 (2-6/09/13 - European Conference on Artificial Life 2013 - PC member
- 2013 (6-10/07/13) - Genetic and Evolutionnary Computation Conference (GECCO) 2013 - PC member
- 2013 (6-7/5/13) - Spatial Computing Workshop 2013 with AAMAS - PC member
- 2013 (13-19/01/13) - Rochebrune 2013 - La preuve et ses moyens - PC member
- 2012 (7-11/07/12) - Generative and Developmental Systems track at GECCO 2012 - PC member
- 2012 (11-16/06/12) - 6th workshop on Dynamic Languages and Applications - PC member
- 2012 (04/06/12) - Spatial Computing Workshop with AAMAS in Valencia/Spain - PC member
- 2011 (03/10/11) - Spatial Computing Workshop with SASO in Ann Arbor (USA) - OC and PC member
- 2011 (08/08/10) - European Conference on Artificial Life - PC member
- 2010 (20/09/10) - 12th International Symposium on Stabilization, Safety, and Security of Distributed Systems (SSS 2010) - PC member of the track Swarm, Amorphous, Spatial, and Complex Systems Track
- 2010 (08/09/10) - ANTS 2010 - Seventh International Conference on Swarm Intelligence - PC member
- 2010 (01/10/10) - Spatial Computing Workshop 2010 with SASO in Budapest (Hungary) - OC and PC member
- 2009 (14/09/09) - Spatial Computing Workshop 2009 with SASO in San Francisco (USA) - OC and PC member
- 2008 (24/11/09) - Agent Based Spatial Simulation at the ISC-PIF in Paris.
- 2008 (20/08/08) - Spatial Computing Workshop 2008 with SASO
- 2009 (19/06/09) - First International Workshop on Morphogenetic Engineering and the organisers page - OC and PC member
- 2008 (06/08/09) - Correlation workshop in Vielsalm and its workgroup
- 2008 (07/07/08) - From Amorphous Computing to Spatial Computing workshop in Paris (France) with G.J. Sussman and C. Teuscher (AI07/ISC/RNSC)
- 2007 (18/07/07) - Amorphous Computing workshop with D. Coore in Paris (France) - OC member
- 2006 (03/09/06) - Dagstuhl Seminar, Computing Media and Languages for Space-Oriented Computation in Dagstuhl (Germnay)
- 2004 (15/09/04) - Unconventional Programming Paradigm in Mont Saint Michel (France) - OC and PC member
Working Groups, Networks...
Nowadays
I'm a member of
- « GDR Internet, IA et Société » more specifically of Axe 3. Un internet inclusif et durable in the subgroup Politique environnementale du numérique lead by C. Marquet and S. Quinton
Previously
I participate in the following working groups and networks
- In computer science
 in the LTP Working Group
in the LTP Working Group
- In synthetic biology and modelling in a broader sense
- ACI Vicanne on Modélisation dynamique et simulation des systèmes biologiques
- In complex systems with the Institute of Complex Systems in Paris Ile de France
 Research network no 54: Groupe de Recherches et d’Études en Biologie Synthétique (headed by F. Delaplace)
Research network no 54: Groupe de Recherches et d’Études en Biologie Synthétique (headed by F. Delaplace) Research project 49 (AI/008/49): Hybrid Modelling of Diffusion-Reactions Processes (with H. Berry and A. Lesne)
Research project 49 (AI/008/49): Hybrid Modelling of Diffusion-Reactions Processes (with H. Berry and A. Lesne)
-
- Proposal for an ANR Domaines Émergents (DEFIS) (access restricted)
I participate in the following projects
- 2004–2007 ACI JC9077 NANOPROG : une approche langage pour le nanocalcul et la simulation des nanosystèmes biologiques with F. Delaplace, F. Gruau, J. Cohen and A. Spicher. scientific report
Previous Research
The 8 1/2 Project
Everything that concerns Otto e mezzo can be found on its own webpage.
The Amalgams
Taking part in the 8 1/2 project, I have defined a formalism based on names an name capture.
Binding and substitution of bound variables are major is- sues in the design of languages. The mechanism of name capture lies at the heart of modular, incremental and object-oriented programming. In this formalism, we address the problem of names and name capture on its own by designing, through the use of three operators, a complete and consistent mechanism of name capture for a declarative framework. We perceive expressions with free names as incomplete expressions which will be dynamically completed by a name capture mechanism. The formalism allows the definition of first-class environments. We apply this mechanism to model distributed incremental program construction and to define an object oriented programming style in a declarative language.
Very little is available on the amalgams in english. A good introduction, while never published, is here.
More can be found here.
Bibliography
A complete bibliography can be found as abibtex file or as a web page.
